library(tidyverse)
library(ggplot2)Lab 6
Statistical tests
Overview
While I encourage you to run any test of your choosing, you’ll likely end up using FIsher’s exact/chi-squared test.
- Chi-squares: https://www.westga.edu/academics/research/vrc/assets/docs/ChiSquareTest_LectureNotes.pdf/
- T-test: https://toltex.imag.fr/TwoTestSPLS/
- Fisher’s exact: https://www.biostat.jhsph.edu/~iruczins/teaching/140.652/supp21.pdf/
We will also practice material learned so far to reinforce some of the code learned so far. This week, we’ll learn how to perform chi-square tests. To do so, I am using Mike Love and Irizarry’s book (recommended for life sciences). You can also visit Mike Love’s github page to use the book online here.
Loading libraries
Loading the file: You need not worry about this chunk. You may simply run it. Or you could also download the mice dataset from the website and read in as a csv file for practice.
dir <- "https://raw.githubusercontent.com/genomicsclass/dagdata/master/inst/extdata/"
url <- paste0(dir, "femaleMiceWeights.csv")
dat <- read.csv(url)Student exercise 1. How many mice are in this dataset? 2. Calculate mean weight of the mice 3. Create a scatter plot (like the one I’ve already created) 4. What is the difference in mean weight by diet of the mice?
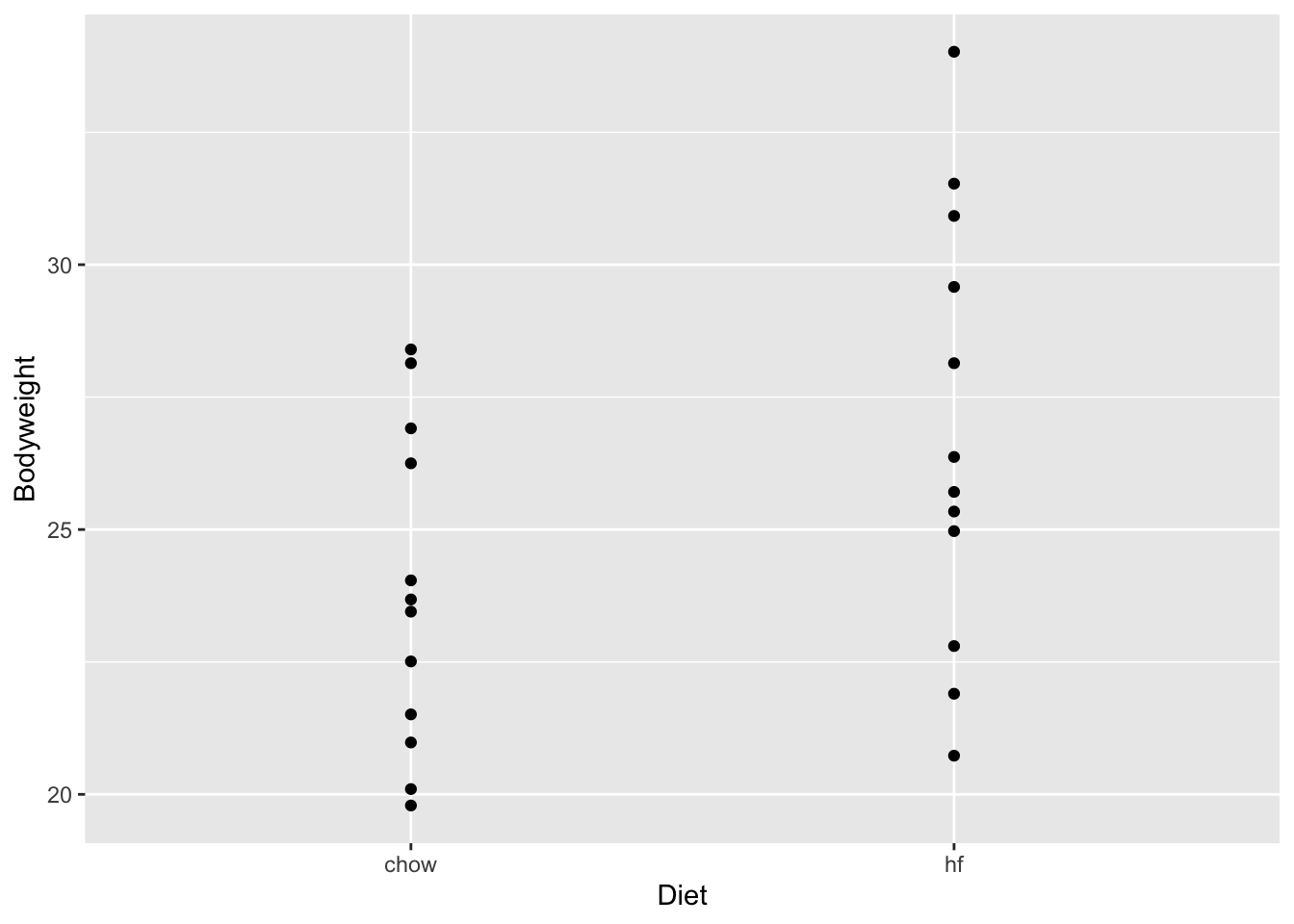
mean_wt_hf
1 3.020833Performing chi-squares and t-tests.
Let’s understand the syntax for both these tests and use p-values to determine significance.
# Perform a t-test for one categorical and one continuous var
t.test(Bodyweight ~ Diet, data = dat)
Welch Two Sample t-test
data: Bodyweight by Diet
t = -2.0552, df = 20.236, p-value = 0.053
alternative hypothesis: true difference in means between group chow and group hf is not equal to 0
95 percent confidence interval:
-6.08463229 0.04296563
sample estimates:
mean in group chow mean in group hf
23.81333 26.83417 # Chi-square requires at least 30 observations, so not ideal in this case
# Also, we do not have two categorical vars so we could recode the weight as high vs low
print(mean(dat$Bodyweight))[1] 25.32375dat_new <-
dat %>%
mutate(new_weight = if_else(Bodyweight > 25.32375, 'High', 'Low'))
test <- chisq.test(table(dat_new$Diet, dat_new$new_weight))
test
Pearson's Chi-squared test with Yates' continuity correction
data: table(dat_new$Diet, dat_new$new_weight)
X-squared = 1.5, df = 1, p-value = 0.2207# Fisher's exact
fisher.test(dat_new$Diet, dat_new$new_weight)
Fisher's Exact Test for Count Data
data: dat_new$Diet and dat_new$new_weight
p-value = 0.2203
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.03289187 1.77138188
sample estimates:
odds ratio
0.2661096 To Do
Nothing!!! This marks the end of our lab section - let’s spend time practicing.